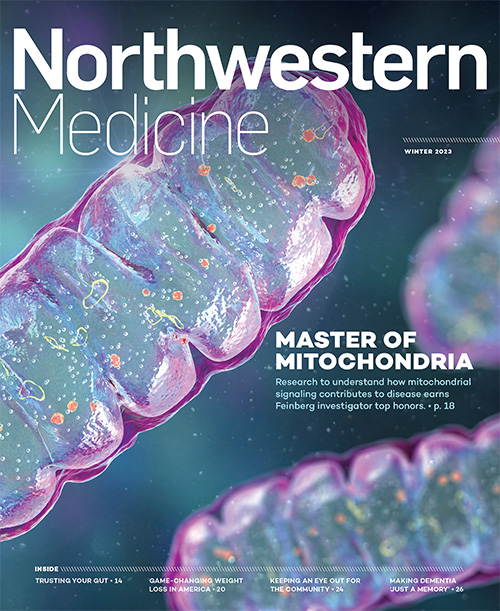
Master of Mitochondria
Navdeep Chandel’s research on mitochondria recognized with the prestigious Lurie Prize in Biomedical Sciences
Trusting Your Gut
Feinberg faculty study how the gut’s microbiome affects a wide range of conditions

Making Dementia ‘Just a Memory’
Robert Vassar, PhD, takes over the Mesulam Center for Cognitive Neurology and Alzheimer’s Disease
Paving the Road to Game-Changing Weight Loss in America
Robert Kushner, MD, links new anti-obesity medications to reducing risk of cardiovascular disease
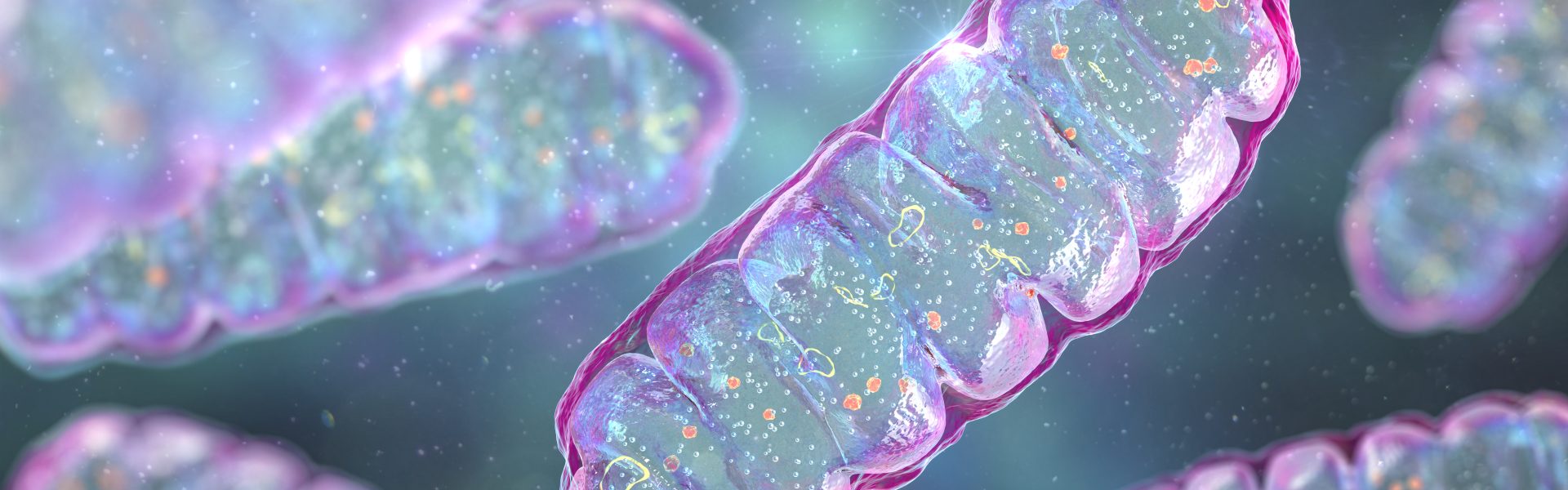
Master of Mitochondria
Navdeep Chandel’s research on mitochondria recognized with the prestigious Lurie Prize in Biomedical Sciences
Trusting Your Gut
Feinberg faculty study how the gut’s microbiome affects a wide range of conditions
Making Dementia ‘Just a Memory’
Robert Vassar, PhD, takes over the Mesulam Center for Cognitive Neurology and Alzheimer’s Disease
Paving the Road to Game-Changing Weight Loss in America
Robert Kushner, MD, links new anti-obesity medications to reducing risk of cardiovascular disease
Research Briefs
New Therapeutic Target for Parkinson’s Disease Discovered
Northwestern Medicine scientists have uncovered a new mechanism by which mutations in a gene, parkin, contribute to familial forms of Parkinson’s disease. The discovery opens…
Pioneering New Methods to Understand Protein Folding
Northwestern Medicine scientists have developed a new technique for measuring protein folding stability on an unprecedented scale, findings detailed in a new study published in…
First Device to Monitor Transplanted Organs, Detect Early Signs of Rejection
Northwestern University scientists have developed the first electronic device for continuously monitoring the health of transplanted organs in real time, as detailed in a study…
Study Discovers Novel Therapeutic Target for Acute Respiratory Distress Syndrome
A new Northwestern Medicine study has discovered a novel therapeutic target and therapeutic agents for older patients with acute respiratory distress syndrome (ARDS), according to…
Alumni News
Alumni Perspective: Paving the Way for Greater Diversity in…
William D. Yates, ‘85 MD, shares his unique experience and perspective as an African American medical student at Feinberg.